Biophysical parameter retrieval from Sentinel-2 images using physics-driven deep learning for PROSAIL inversion
The results presented here are based on published work: Y. Zérah, S. Valero, and J. Inglada. « Physics-constrained deep learning for biophysical parameter retrieval from sentinel-2 images: Inversion of the prosail model« , in Remote Sensing of Environment, doi: 10.1016/j.rse.2024.114309.
This work is part of the PhD of Yoël Zérah, supervised by Jordi Inglada and Silvia Valero. A simpler version of this post, in French, is available on CNES Data Campus blog
Introduction: variable estimation
Remote sensing enables to acquire information about the Earth surface at large scale. Key information that users of remote sensing data are interested in include the nature and properties of the ground elements. Different remote sensors (optical, radar, laser, etc.) enable to characterize different and complementary aspects of the ground.
The Multi-Spectral Instrument (MSI) onboard the Sentinel-2 (S2) satellites captures optical images with up to 10m spatial resolution with a frequency of at least every 5 days. Its 13 spectral channels are especially suited for observing vegetation because they operate in the visible and near infra-red part of the spectrum.
However, beyond direct visual interpretation of optical images, the relevant information is hidden within the acquired signal. Optical imagery, like any other remote sensing data, is first and foremost, a measurement of incident electromagnetic radiation. The raw light measurement is rarely the final objective, e.g. on its own a reflectance level of 0.4153 on the B4 band of the MSI rarely provides useful insight. Depending on the application, different representations of the Earth surface are needed: land cover/use, changes, soil properties, vegetation characteristics, etc. Here, we focus on retrieving vegetation bio-physical variables, e.g. leaf pigments such as chlorophyll and canopy characteristics such as the leaf area index (LAI).
Retrieving actual information of interest usually involves finding relationships between the ground properties and the received signal, i.e. building scientific models. These models are functions of the data that output a prediction of the quantity of interest. Such models are typically found empirically, i.e. built from available data. In recent years, the rise of machine learning (ML) methods and tools, computing power and amount of remote sensing data have enabled to build more powerful models. In particular, deep learning (DL), based on deep neural networks, have emerged as a very powerful framework for learning a variety of tasks.
Performing variable estimation at large scale imposes computational efficiency on proposed methods. For instance, powerful Bayesian estimation approaches such as Monte-Carlo Markov Chain (MCMC) are prohibitively costly to run. On the other hand, once trained, DL approaches are relatively efficient and can be accelerated using dedicated hardware.
Continuous variables such as bio-physical variables are estimated using regression models. The typical approach is to train a ML model (neural networks, random forests, Gaussian processes, etc.) using samples of pairs of reference bio-physical variables (e.g. the LAI) with matching remote sensing observation (e.g. S2 image). This is supervised training, where a label (the desired output) is provided for each training input data. A key issue with this approach is that there is usually not enough reference bio-physical data to build labelled data-sets. Reference bio-physical variables must be measured on the ground which makes it expensive and labor-intensive. Available reference data is insufficient for both characterizing the diversity in vegetation, and for optimizing models with many parameters such as neural networks.
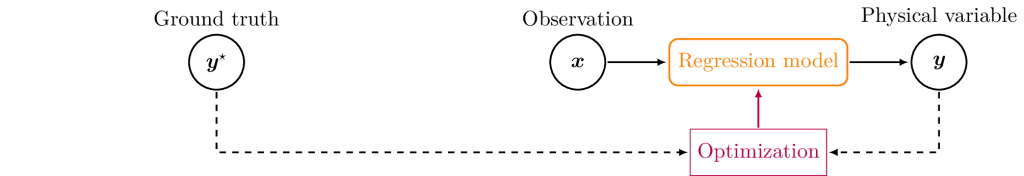
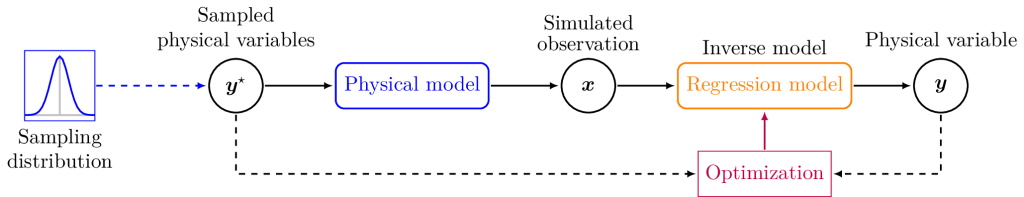
This is why training data is commonly simulated rather than measured. An observational model simulates a remote sensing observation from a set of bio-physical variables, and enables to generate training samples. The regression model is then trained using simulations, and its performances are validated using the few reference data available. This regression model de facto performs the inversion of the observational model.
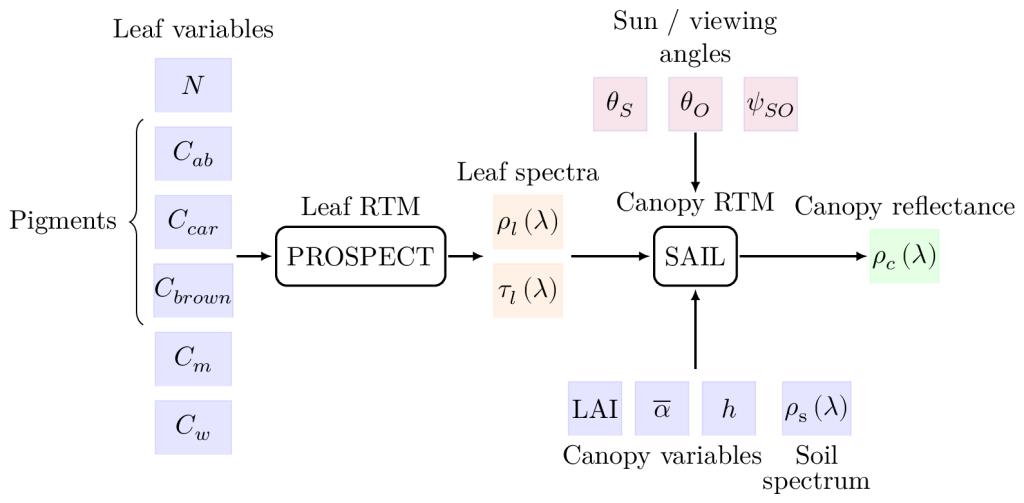
Physical model : PROSAIL
PROSAIL is a popular and efficient radiative transfer model (RTM) that has been successfully used to simulate satellite reflectance measurements from randomly sampled bio-physical variables. It is the composite model of the PROSPECT RTM, which simulates the optical properties of leaves from leaf variables (e.g. pigment concentrations), and the SAIL RTM which simulates canopy reflectances from canopy variable and leaf reflectance. Several version of PROSPECT and SAIL enable to take into account different properties of the vegetation canopy into account. Finally, the spectral sensitivity of an instrument (e.g. the MSI) enables to estimate bottom-of-atmosphere spectral band measurements (e.g. S2 bands).
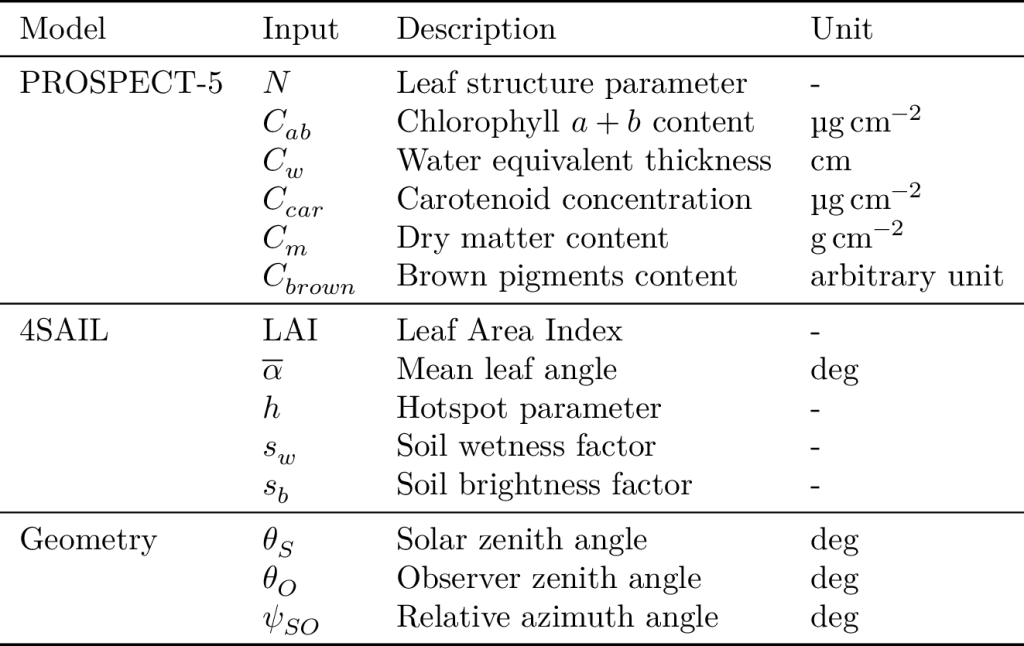
The also popular Biophysical Processor of the Sentinel Application Platform (SNAP) is based on a 2-layer regression neural network which is trained using PROSAIL simulations. Specifically, a neural network is dedicated to predict each of five bio-physical variables, among which are the LAI and the canopy chlorophyll content (CCC). A key aspect of SNAP is the distribution chosen for sampling the input bio-physical variables for PROSAIL to generate simulations.
It has been shown that this distribution is paramount to the performance of the trained neural network. Unfortunately, choosing this distribution is difficult because:
- it is necessary to sample jointly all PROSAIL variables because of the range of combinations is limited in nature,
- the lack of available in-situ reference data prevents an accurate distribution estimation, and some parameters are not commonly measured (e.g. leaf inclination angle distribution (LIDF), hot-spot parameter, soil spectrum).
Furthermore, the tedious work of finding the right sampling distribution must be started over each time either the simulator, or the target sensor changes.
As such, there is potential in unsupervised approaches that do not require a labeled data-set, i.e. a method that estimates bio-physical variables from remote sensing observations without requiring a reference, which is currently either too scarce with ground measurement or dependent on an arbitrary distribution with simulations. Therefore in the following is presented our DL approach, which can be trained directy on L2A Sentinel-2 images to estimate bio-physical variables, using PROSAIL as a physical bias.
Data-sets
This study requires two kinds of data-sets:
- A training data-set, for tuning our DL model,
- An evaluation data-set for assessing its performance on bio-physical variable retrieval.
Training patch data-set
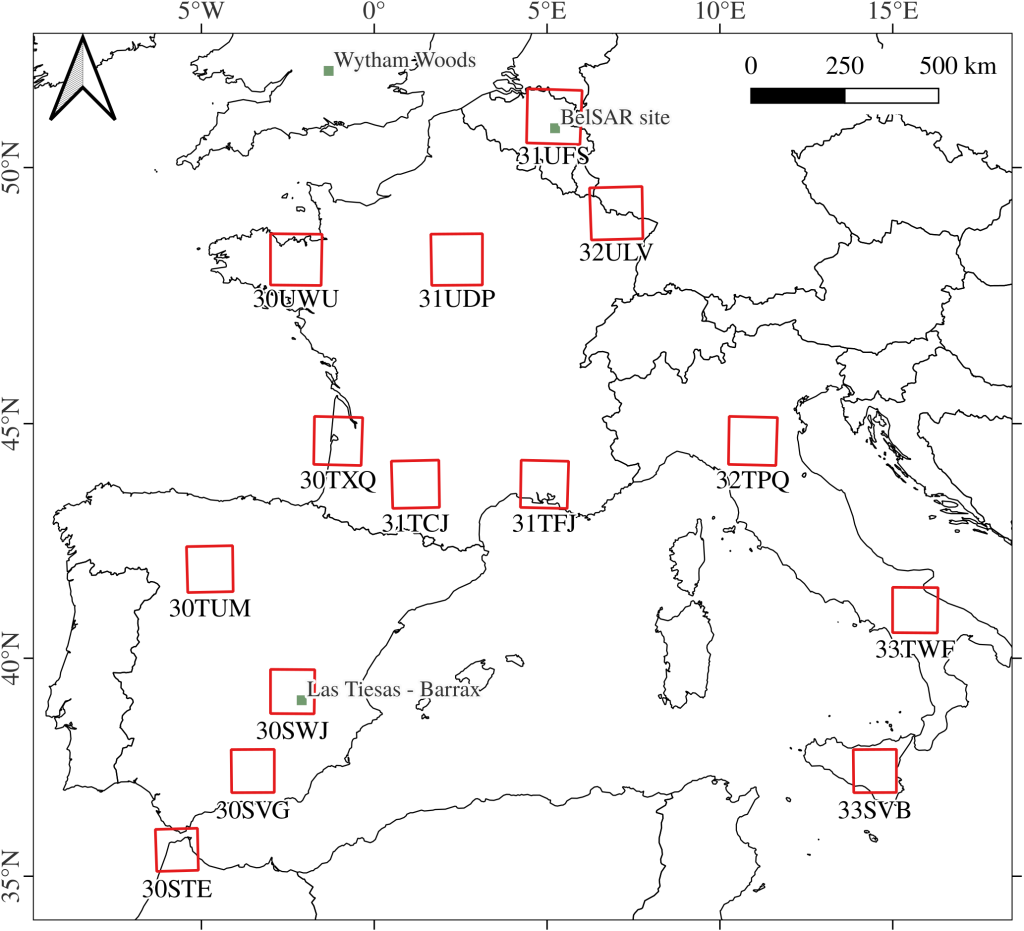
The training data-set is made of a collection L2A S2 image patches from the THEIA catalog. These patches were extracted from 20 5120m × 5120m regions of interest (ROI) in 14 S2 tiles over western Europe to provide a variety of landscapes. These ROI are centered on vegetated areas, either crops or forests, although non-vegetation elements may be present. For each ROI, multiple acquisitions between 2015 and 2019 were collected to account for temporal variability. Patches with detected clouds were filtered out. In total, this training data contains over 24 million pixels.
Testing data-set
A set of patches from each ROI at each acquisition dates are set aside for computing certain certain statistics, as will be discussed below. These patches do not intervene during the training of our model.
Evaluation data-set
The evaluation data-set contains 211 LAI and 121 CCC measurements of crops from 4 campaigns at 3 different test sites:
- 3 FRM4Veg campaigns in Las Tiesas – Barrax (Spain) in 2018 and 2021 and in Wytham Woods (England) in 2018,
- The BelSAR campaign (Belgium) in 2018.
The measurements were associated with S2 acquisitions of the test sites. Most measurement weren’t performed on the same day of a valid S2 acquisitions: for those cases, the first valid image before and after the measurement were collected. This way, predictions of biophysical variables can be interpolated to the day of measurement and compared to the reference values.
While the BelSAR and Barrax sites are located within the tiles T30SWJ and T31UFS which were selected for the training data-set, we ensured that the testing sites were not part of the chosen ROI, so that there are no pixel in common between the training data-set and the evaluation data-set.
Method: Self-supervised deep learning for biophysical parameter estimation
Variationnal autoencoder
The approach proposed here is based on representation learning with variational autoencoders (VAE). VAE are a popular type of deep learning models that were developed from variational inference. A VAE is composed of:
- a neural network encoder that takes data x as input and produces as output an estimate of the posterior distribution of latent variables (or latent distribution),
- a neural network decoder that takes samples z of the latent distributions as input, and produces a likelihood estimate of the encoder’s input data x, i.e. a distribution whose samples arereconstructions.
VAE are trained with the so-called ELBO objective function which promotes closeness between the latent distribution and a prior distribution, and penalizes the log-likelihood of the decoder’s output w.r.t. the input data x (also called reconstruction loss). VAE are self-supervised: no label or reference data is required, since the model target is the input data x itself.
The latent distribution is a representation of the input data, that promotes some statistical properties that can be useful for other tasks. Unfortunately, this is not helpful when trying to estimate specific physical quantities, because the latent variables are typically not interpretable: they are an abstract decomposition of the input data that is not constrained to match any specific physical concept.

PROSAIL-VAE
Our approach proposes to constrain the latent variables so that they specifically match physical variables, by integrating physical bias. PROSAIL-VAE is defined as a VAE that takes S2 optical images as input and in which the neural network decoder is replaced by PROSAIL. The output of the decoder (i.e. the reconstructions) are PROSAIL simulations generated from the latent samples, that are semantically tied to PROSAIL input variables. When PROSAIL-VAE is trained, the objective function ensures that the reconstructions matches the input optical image, and because they are generated with a physical model, the latent variables are forced to take values that match their PROSAIL input counterparts.
In PROSAIL-VAE, contrary to classical VAE, only the encoder is trained. When the training is completed, the PROSAIL decoder is discarded, and the encoder can be used as a full probabilistic model inverse of PROSAIL: all PROSAIL variables are estimated simultaneously. Crucially, PROSAIL-VAE can be trained directly using S2 images, without providing reference bio-physical variables.
It is important to note that the output of the encoder in PROSAIL-VAE are distribution parameters (like in classical VAE), i.e. the posterior distribution of PROSAIL parameters is predicted. This enables to perform uncertainty quantification.
Key aspects of the method, such as:
- the Monte-Carlo reconstruction loss, that enables to estimate a likelihood distribution using a deterministic model,
- the need for a differentiable physical model in the decoder,
- considerations on the variational distribution and the prior distribution and
- uncertainty quantification
are discussed in the original RSE article, along with our previous IEEE-TGRS article on similar work, and at length in Yoël Zérah’s thesis.

Results
The following present the results obtained with a PROSAIL-VAE model that was trained on the above-described training patch data-set, i.e. the estimation of PROSAIL input variables using the encoder of PROSAIL-VAE. Estimates of bio-physical variables are the expected values of the distributions produced by PROSAIL-VAE.
Validation of LAI and CCC estimation
The evaluation data-set was used to validate the prediction of PROSAIL-VAE from S2 images on measured LAI and CCC reference data. SNAP predictions on the same data-set were used as baseline for comparison. To assess the parameter retrieval performance, regression metrics such as the R² score and the RMSE were computed.
For the LAI, the performance of PROSAIL-VAE and SNAP are very similar, and the predictions of both models are also very similar for each sample. For instance, LAI of poppy seeds samples were underestimated for both models. While PROSAIL-VAE has better metrics, it exhibits a small positive bias for low LAI values.

For the CCC, SNAP directly outputs a parameter estimate. However, it is estimated in PROSAIL-VAE as the product CCC = LAI × C_ab, with C_ab the leaf chlorophyll content, which makes the CCC prediction reliant on the accuracy of retrieval of two parameters. Despite this, PROSAIL-VAE clearly outperforms SNAP, which tends to overestimate this parameter.

Biophysical parameters retrieved from deciduous forests are predicted within a smaller range than measured in situ, for both LAI and CCC, and for both model. We explain such a behavior by the fact that PROSAIL which is used (differently) in both approaches better suited to simulate crops than forests, for which other RTM are better.
Assessing the prediction of other PROSAIL parameters
For other parameters, it is more difficult to assess whether they are well estimated by PROSAIL-VAE, because the used evaluation data-set doesn’t contain reference measurements for them. Using only this data, regression metrics cannot be estimated.
To qualitatively assess the retrieval of these parameters, we derived their empirical distributions and pairwise correlations. Specifically, performed PROSAIL variable retrieval with PROSAIL-VAE on the images of the testing data-set, and gathered the predictions to derive an aggregate distribution for all parameters.
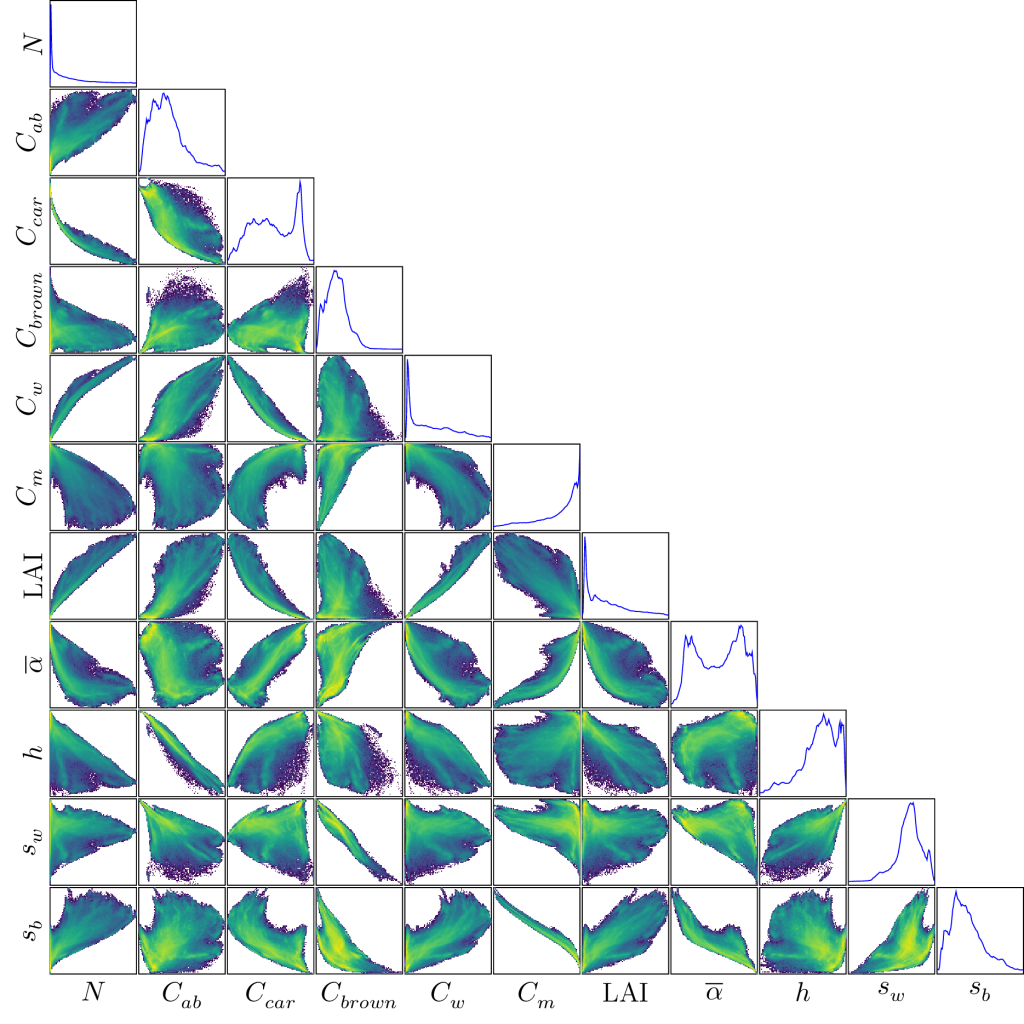
This enables to assess that the retrieval performances vary among parameters. For instance the LAI distribution is consistant with log-normal distributions observed in measurement campaigns. On the other hand, some retrieved parameters show some issues. The carotenoid content C_car exhibits an negative correlation with the chlorophyll content C_ab, when it should be positively correlated instead. This parameter is notorious for being difficult to estimate, especially with Sentinel-2 whose bands are barely sensitive to its spectral signature. The range of certain retrieved parameter is too restricted to be realistic, such as the leaf structure parameter N, whose values are concentrated near its lower bound 1.2.
This can be also explained by the fact that this inversion problem is ill-posed, with more parameters to estimate than input spectral bands.
Conclusion and perspectives
Despite the good results achieved by PROSAIL-VAE on our evaluation data, the goal of this work was never to establish a new retrieval method that performed better than the operational SNAP, that has been extensively validated.
However, we showed that a different way for inverting models and retrieving variables was possible, one that isn’t supervised on synthetic data-sets, that are painstaklingly designed to maximize performance. Our PROSAIL-VAE approach is trained directly on S2 images, effectively changing the difficult choice of parameter sampling distribution to an arguably easier choice of images. Additionnally, all PROSAIL parameters are retrieved at once by a single model, even if for now they are not all well estimated.
This approach offers many potential future developments.
- The parameter retrieval could be improved by enforcing correlation constraints during training,
- The approach is not specific to PROSAIL, so it could be swapped for other models (such as the INFORM RTM, better suited for forests).
- All physical models, including PROSAIL, have assumptions that are never perfect: the residuals of the simulations could be estimated using a complementary neural network decoder.
- The approach could enable estimating parameters by using the data of different sensors at once (e.g. optical such as S2 and SAR such as S1), by feeding different data to the encoder, and by using multiple physical models as decoders to reconstruct the input data and train the encoder.
Some of these perspectives will be explored during the PhD of Kevin De Sousa which started last year at CESBIO.
Auteur : Yoel Zerah
Acknowledgments
This work was supported by the Natural Intelligence Toulouse Institute (ANITI) from the Université
Fédérale Toulouse Midi-Pyrénées under Grant Agreement ANITI ANR-19-P3IA-0004, by the ANR-JCJC DeepChange project under Grant Agreement 20-CE23-0003, by the ANR MAESTRIA project under the grant agreement 18-CE23-0023 and by the Centre national d’études spatiales (CNES) under Grant Agreement n° 51/19560.
We would like to thank CNES for the provision of its high performance computing (HPC) infrastructure to run the experiments and the associated help.